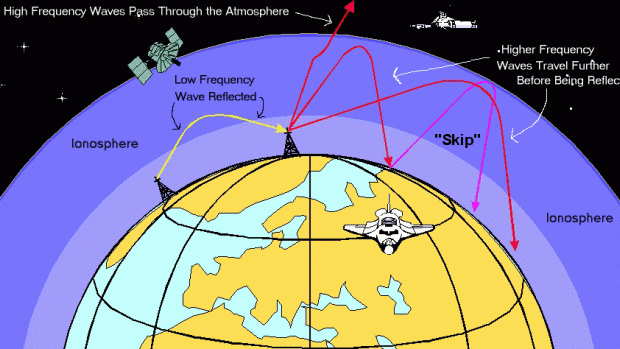
- Pictured: Ionospheric effects on radio waves at different frequencies.
- Source: NASA https://www.swpc.noaa.gov/phenomena/ionosphere
This research project was undertaken to fulfill requirements for 'Project 1' of City College of San Francisco's MATH 108 Fundamentals of Data Science course.
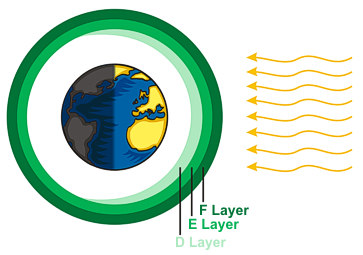
Earth's atmosphere is composed of several layers, each with different physical properties. The ionosphere is region of the atmosphere containing several sub-layers of electrically charged (ionized) gas particles. Depending on the density and energy of these gas particles, radio frequency (RF) traveling through these layers can be absorbed, reflected back to earth, or lost to space.

Sub-layers tend to follow diurnal (day/night) patterns, with E & F being persistent, and D, F1 & F2 emerging only during the day.
Methods of measuring the properties (sounding) the ionosphere range from active radar-like radio frequency probes to passive GPS satellite signal observation. Ionosonde is the term used for active radio frequency probes.
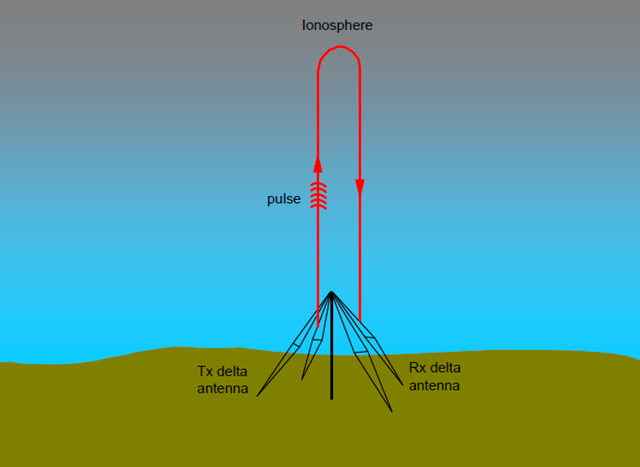
Ionosonde data can be represented visually in the form of an ionogram. Ionograms typically include data such as the Frequency of F2, Frequency of E, Total Electron Content, et al.
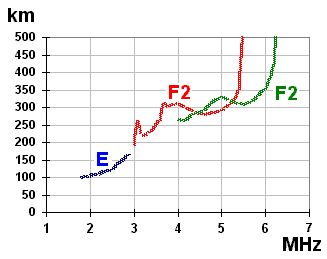
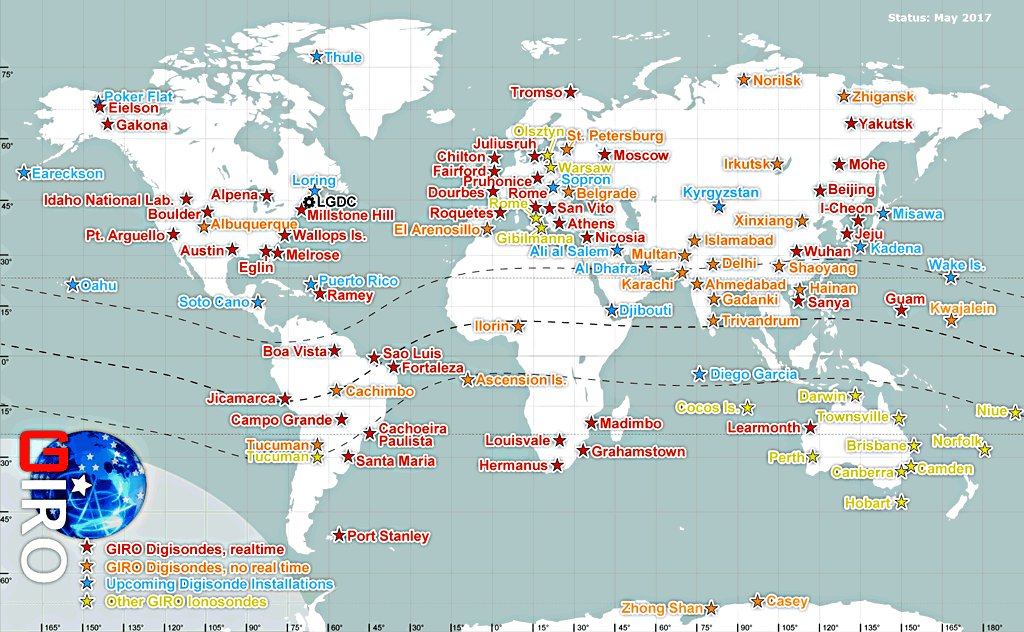
An international consortium of research institutions runs several ionospheric sounding stations around the planet. These stations probe the ionosphere periodically through the day, year round. This data is used to predict ionospheric conditions that may effect long range radio transmission, satellite signal reception, and global navigation satellite positioning.
Heliophysics has the strongest influence on ionospheric conditions, including day/night, solar wind, sun cycles, sun spots, coronal mass ejections (CMEs), et al. Earth weather & geophysics also influence the ionosphere, including weather & storms, tsunamis & earthquakes.
This project aims to use the global ionospheric surveillance network dataset to determine if the Hunga Tonga volcanic eruption in January 2022 had a measurable effect on the ionosphere.
Variables & measurements we intend to measure include:
MHz.10^16 electrons/m² = 1 TECUPotential limitations to the data used in this project include:
This project originally used raw MIDS data from NOAA, but switched over to use FastChar data from GIRO. Consider all MIDS-related functions DEPRECATED in favor of FastChar.
The data used in this project comes from these sources:
Ionosonde MIDS & FastChar:
"A large eruption commenced on 15 January 2022 at 04:20 local time, sending clouds of ash 20 km (12 mi) into the atmosphere."
https://volcano.si.edu/ShowReport.cfm?doi=10.5479/si.GVP.WVAR20220112-243040
https://cimss.ssec.wisc.edu/satellite-blog/archives/44214
https://en.wikipedia.org/wiki/2022_Hunga_Tonga_eruption_and_tsunami
Look at Guam barometric pressure data to see when pressure wave hit:
https://www.ndbc.noaa.gov/station_history.php?station=aprp7# Python standard modules
import datetime
import os
import time
import urllib.request
# CCSF MATH 108 / UCB DATA 8 imports
from datascience import *
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plots
plots.style.use('fivethirtyeight')
# Other external modules used in this notebook
import pandas as pd
# Into which directories would we save our data:
FASTCHAR_DIR = 'fastchar'
CWOP_DIR = 'cwop'
# If set to True, the following will flush existing data:
FLUSH_FASTCHAR = True
FLUSH_CWOP = False
# Which measurements do we want to retrieve from FastChar?
FASTCHAR_NAMES = 'foF2,foF1,foE,foEs,MUFD,fmin,TEC'
# Hunga Tonga Volcano location:
HUNGA_TONGA_LAT = 20.536
HUNGA_TONGA_LON = 175.382
# When did the eruption start (UTC) January 15, 04:20 UTC
ERUPTION_START = datetime.datetime(2022, 1, 15, 4, 20, tzinfo=datetime.timezone.utc)
# To access this servlet use format:
# DIDBGetValues?ursiCode=CODE&charName=NAME&fromDate=yyyy.mm.dd%20(...)%20hh:mm:ss&toDate=yyyy.mm.dd%20(...)%20hh:mm:ss
# For example: DIDBGetValues?ursiCode=DB049&charName=foF2&fromDate=2007.06.25%20(...)%2000:00:00&toDate=2007.06.26%20(...)%2000:00:00
# or: DIDBGetValues?ursiCode=DB049&charName=foF2&fromDate=2007.06.25&toDate=2007.06.26
def get_fastchar(usri_code: str) -> str:
"""Downloads FastChar data from the GIRO FastChar API."""
base_url = 'https://lgdc.uml.edu/common/DIDBGetValues'
from_date = '2022.01.01'
to_date = '2022.01.31'
dmuf = 3000
url = (f"{base_url}?ursiCode={usri_code}&charName={FASTCHAR_NAMES}"
f"&fromDate={from_date}&toDate={to_date}&DMUF={dmuf}")
print(f"Getting FastChar from {url}")
file_name: str = f"{usri_code}.txt"
file_path: str = f"{FASTCHAR_DIR}/{file_name}"
# Don't download again
if os.path.exists(file_path) and not FLUSH_FASTCHAR:
print(f"FastChar exists: {file_path}")
return
try:
with urllib.request.urlopen(url) as mids_df:
with open(file_path, "wb+") as mids_file:
print(f"Saving FastChar to {file_path}")
mids_file.write(mids_df.read())
return file_path
except Exception as exc:
print(f"Exception: {exc}")
return
def calc_dist(lat: float, lon: float) -> float:
"""Calculates distance from Hunga Tonga Volcano."""
func = haversine_distance
return func(HUNGA_TONGA_LAT, HUNGA_TONGA_LON, lat, lon)
def fix_ts(ts: str) -> datetime.datetime:
"""Parses the given timestamp as a datetime.datetime in UTC."""
return datetime.datetime.fromisoformat(f"{ts}+00:00")
def format_fastchar_vals(val):
"""Replaces empty readings (---) with 0 and casts to float."""
if not isinstance(val, (float, int)):
return float(val.replace('---', '0'))
else:
return val
def download_sleep(inc: int = 20) -> None:
"""Sleeps for a random period to avoid flooding GIRO API."""
sleep_time = np.random.random() * inc
print(f"Sleeping for {sleep_time}")
time.sleep(sleep_time)
def get_cwop(site: str) -> None:
"""Downloads CWOP data from the weather data archive."""
url: str = f"https://weather.gladstonefamily.net/cgi-bin/wxobservations.pl?days=56&site={site}"
file_name: str = f"{site}.csv"
file_path: str = f"{CWOP_DIR}/{file_name}"
# Don't download again
if os.path.exists(file_path) and not FLUSH_MIDS:
print(f"CWOP exists: {file_path}")
return
print(f"Getting CWOP from {url}")
try:
with urllib.request.urlopen(url) as mids_df:
with open(file_path, "wb+") as mids_file:
print(f"Saving CWOP to {file_path}")
mids_file.write(mids_df.read())
return file_path
except Exception as exc:
print(f"Exception: {exc}")
return
# Cargo-culted radian distance calculation:
def haversine_distance(lat1, lon1, lat2, lon2):
"""
https://towardsdatascience.com/heres-how-to-calculate-distance-between-2-geolocations-in-python-93ecab5bbba4
"""
# Earth radius:
# Miles: 3959.87433
# km: 6372.8
r = 6372.8
phi1 = np.radians(lat1)
phi2 = np.radians(lat2)
delta_phi = np.radians(lat2 - lat1)
delta_lambda = np.radians(lon2 - lon1)
a = np.sin(delta_phi / 2)**2 + np.cos(phi1) * \
np.cos(phi2) * np.sin(delta_lambda / 2)**2
res = r * (2 * np.arctan2(np.sqrt(a), np.sqrt(1 - a)))
return np.round(res, 2)
station_names = make_array(
'Pt. Arguello',
'Wallops Is.',
'Guam',
'Learmonth',
'Boulder',
'Idaho National Lab'
)
usri_codes = make_array(
'PA836',
'WP937',
'GU513',
'LM42B',
'BC840',
'IF843'
)
lats = make_array(
34.80,
37.90,
13.62,
-21.80,
40.00,
43.81
)
lons = make_array(
239.50,
284.50,
144.86,
114.10,
254.70,
247.32
)
station_names = Table().with_columns(
'Name', station_names,
'USRI', usri_codes,
)
station_locations = Table().with_columns(
'USRI', usri_codes,
'lat', lats,
'lon', lons
)
stations = station_names.join('USRI', station_locations, 'USRI')
stations
| USRI | Name | lat | lon |
|---|---|---|---|
| BC840 | Boulder | 40 | 254.7 |
| GU513 | Guam | 13.62 | 144.86 |
| IF843 | Idaho National Lab | 43.81 | 247.32 |
| LM42B | Learmonth | -21.8 | 114.1 |
| PA836 | Pt. Arguello | 34.8 | 239.5 |
| WP937 | Wallops Is. | 37.9 | 284.5 |
def download_data():
"""Downloads the ionosonde data."""
if not os.path.exists(FASTCHAR_DIR):
os.mkdir(FASTCHAR_DIR)
for usri in stations.column('USRI'):
print(f"Downloading ionosonde data for {usri=}")
if get_fastchar(usri):
download_sleep()
# Only need to run once to download ionosonde data:
# download_data()
def populate_dt():
"""Populates the ionosonde DataTables."""
main_dt = Table()
for usri in stations.column('USRI'):
print(f"Populating DataTable for {usri=}")
# Calculate the distance from Hunga Tonga to the site:
distance = calc_dist(
stations.where('USRI', usri).column('lat').item(0),
stations.where('USRI', usri).column('lon').item(0)
)
file_name: str = f"{usri}.txt"
file_path: str = f"{FASTCHAR_DIR}/{file_name}"
# Read the 'fixed width' data as a pandas DataFrame,
# then convert to a DataTable.
station_df = pd.read_csv(
file_path,
sep='\s+',
skiprows=np.arange(0, 26)
)
station_dt = Table().from_df(station_df).relabel('#Time', 'Time')
# Fix the values for some of our columns:
station_dt = station_dt\
.where('foF2', are.not_containing('---'))\
.where('TEC', are.not_containing('---'))
station_dt = Table().with_columns(
'ts', station_dt.apply(
lambda t: datetime.datetime.fromisoformat(t.replace('Z', '+00:00')), 'Time'),
'foF1', station_dt.apply(format_fastchar_vals, 'foF1'),
'foF2', station_dt.apply(float, 'foF2'),
'foE', station_dt.apply(format_fastchar_vals, 'foE'),
'MUFD', station_dt.apply(format_fastchar_vals, 'MUFD'),
'fmin', station_dt.apply(format_fastchar_vals, 'fmin'),
'TEC', station_dt.apply(float, 'TEC'),
'CS', station_dt.apply(int, 'CS')
)
station_dt['USRI'] = usri
station_dt['Distance'] = float(distance)
# Append/join this DataTable to the existing DataTable:
if not main_dt:
main_dt = station_dt
else:
main_dt.append(station_dt)
# At this point our DataTable should be sanitized.
# Return it sorted by Time Stamp.
return main_dt.sort('ts')
# Populate our DataTable and give us a preview:
main_dt = populate_dt()
main_dt
Populating DataTable for usri='BC840' Populating DataTable for usri='GU513' Populating DataTable for usri='IF843' Populating DataTable for usri='LM42B' Populating DataTable for usri='PA836' Populating DataTable for usri='WP937'
| ts | foF1 | foF2 | foE | MUFD | fmin | TEC | CS | USRI | Distance |
|---|---|---|---|---|---|---|---|---|---|
| 2022-01-01 00:00:00+00:00 | 4.45 | 9.413 | 3.38 | 28.16 | 2 | 26.3 | 85 | GU513 | 3329.3 |
| 2022-01-01 00:00:00+00:00 | 0 | 4.325 | 0 | 15.08 | 1.65 | 3.9 | 80 | IF843 | 7018.86 |
| 2022-01-01 00:00:00+00:00 | 0 | 6.1 | 0 | 24.47 | 2.38 | 3.4 | 60 | PA836 | 6406.29 |
| 2022-01-01 00:00:08+00:00 | 0 | 3.6 | 0 | 11.53 | 1.7 | 2.6 | 100 | WP937 | 10179.4 |
| 2022-01-01 00:05:08+00:00 | 0 | 3.6 | 0 | 11.74 | 1.7 | 1.8 | 100 | WP937 | 10179.4 |
| 2022-01-01 00:07:30+00:00 | 0 | 9.313 | 3.4 | 27.38 | 2 | 27.3 | 95 | GU513 | 3329.3 |
| 2022-01-01 00:07:30+00:00 | 0 | 5.775 | 0 | 14.99 | 3.2 | 12 | 90 | PA836 | 6406.29 |
| 2022-01-01 00:10:08+00:00 | 0 | 3.5 | 0 | 11.74 | 1.7 | 1.6 | 100 | WP937 | 10179.4 |
| 2022-01-01 00:15:00+00:00 | 4.7 | 9.238 | 3.4 | 27.27 | 2 | 28.9 | 85 | GU513 | 3329.3 |
| 2022-01-01 00:15:00+00:00 | 0 | 4.275 | 0 | 14.18 | 1.75 | 3.7 | 70 | IF843 | 7018.86 |
... (30092 rows omitted)
We're going to focus our investigation on the station at Guam.
# Select just one station to focus on:
focus_dt = main_dt.where('USRI', 'GU513')
For our zoomed-out view we'll look at data for the entire month of January. Red dot represents the eruption.
We'll look at two values from the ionosonde, the foF2 and the total electron content (TEC). There is an association between these two values - that is: As the TEC increases so does the foF2.
focus_dt.scatter('foF2', 'TEC')
The plot below shows TEC measurements for all of January, which seem to follow some diurnal patterns, climaxing during the day and dropping at night.
plots.clf()
plots.figure(figsize = (20, 10))
plots.plot(
focus_dt.column('ts'),
focus_dt.column('TEC'),
label='TECU'
)
plots.ylabel('Total Electron Content (10^16 m^-2)')
plots.scatter(x=ERUPTION_START, y=np.min(focus_dt.column('TEC')), color='red', zorder=100)
plots.legend()
plots.show()
<Figure size 432x288 with 0 Axes>
To 'zoom-in' we're going to take a snapshot of our data within the day of the eruption.
# Concentrate on a period surrounding the volcanic eruption:
event_start = datetime.datetime(2022, 1, 15, tzinfo=datetime.timezone.utc)
event_end = datetime.datetime(2022, 1, 16, tzinfo=datetime.timezone.utc)
snap_dt = focus_dt.where('ts', are.between(event_start, event_end))
snap_dt
| ts | foF1 | foF2 | foE | MUFD | fmin | TEC | CS | USRI | Distance |
|---|---|---|---|---|---|---|---|---|---|
| 2022-01-15 00:00:00+00:00 | 4.7 | 7.563 | 3.2 | 27.93 | 2 | 11.8 | 95 | GU513 | 3329.3 |
| 2022-01-15 00:07:30+00:00 | 4.47 | 7.313 | 3.25 | 25.1 | 2 | 17.6 | 90 | GU513 | 3329.3 |
| 2022-01-15 00:15:00+00:00 | 0 | 7.375 | 3.25 | 23.61 | 2 | 17.1 | 95 | GU513 | 3329.3 |
| 2022-01-15 00:22:30+00:00 | 0 | 7.55 | 3.3 | 24.85 | 2.03 | 16.2 | 90 | GU513 | 3329.3 |
| 2022-01-15 00:30:00+00:00 | 5 | 7.688 | 3.3 | 24.87 | 2.03 | 19.2 | 95 | GU513 | 3329.3 |
| 2022-01-15 00:37:30+00:00 | 5.1 | 7.863 | 3.35 | 24.82 | 2 | 20.5 | 95 | GU513 | 3329.3 |
| 2022-01-15 00:45:00+00:00 | 0 | 8.1 | 3.45 | 25.24 | 2.5 | 20.8 | 95 | GU513 | 3329.3 |
| 2022-01-15 00:52:30+00:00 | 3.95 | 8.388 | 3.33 | 26.02 | 2.5 | 21.7 | 95 | GU513 | 3329.3 |
| 2022-01-15 01:00:00+00:00 | 5.3 | 8.613 | 3.35 | 26.78 | 2.38 | 21.9 | 95 | GU513 | 3329.3 |
| 2022-01-15 01:07:30+00:00 | 0 | 8.8 | 3.53 | 26.84 | 2.5 | 23.8 | 95 | GU513 | 3329.3 |
... (134 rows omitted)
This plot shows the typical diurnal pattern of daytime peak and nightime low, but with a considerable spike shortly after the eruption.
plots.clf()
plots.figure(figsize = (20, 10))
plots.plot(
snap_dt.column('ts'),
snap_dt.column('TEC'),
label='TECU'
)
plots.ylabel('Total Electron Content (10^16 m^-2)')
plots.xlabel('Time (UTC)')
plots.scatter(x=ERUPTION_START, y=np.min(snap_dt.column('TEC')), color='red', zorder=100)
plots.legend()
plots.show()
<Figure size 432x288 with 0 Axes>
Similar to the TEC pattern plotted above, the foF2 plot shows usual diurnal pattern, with a notable peak-trough-peak shortly after the eruption, followed by another peak later in the day.
plots.clf()
plots.figure(figsize = (20, 10))
plots.plot(
snap_dt.column('ts'),
snap_dt.column('foF2'),
label='foF2'
)
plots.ylabel('Frequency of F2 (MHz)')
plots.xlabel('Time (UTC)')
plots.yticks(np.arange(2, 15, step=0.5))
plots.scatter(x=ERUPTION_START, y=np.min(snap_dt.column('foF2')), color='red', zorder=100)
plots.legend()
plots.show()
<Figure size 432x288 with 0 Axes>
This histogram of foF2 shows us the distribution of foF2 readings throughout our snapshot period, with two prominent areas around 2.5 and 10.75 MHz.
snap_fof2 = snap_dt.column('foF2')
snap_bins = np.arange(np.min(snap_fof2), np.max(snap_fof2)+1, .25)
snap_dt.hist('foF2', bins=snap_bins)
# Create a new DataTable of the day before the eruption
day_before = datetime.timedelta(hours=-24)
day_before_dt = focus_dt.where(
'ts',
are.between(event_start + day_before, event_end + day_before)
)
day_before_dt
| ts | foF1 | foF2 | foE | MUFD | fmin | TEC | CS | USRI | Distance |
|---|---|---|---|---|---|---|---|---|---|
| 2022-01-14 00:00:00+00:00 | 0 | 10.275 | 3.42 | 33.05 | 2.05 | 23.6 | 95 | GU513 | 3329.3 |
| 2022-01-14 00:07:30+00:00 | 0 | 10.525 | 3.45 | 33.49 | 2 | 20.4 | 95 | GU513 | 3329.3 |
| 2022-01-14 00:15:00+00:00 | 0 | 10.725 | 3.38 | 33.62 | 2.08 | 30.2 | 95 | GU513 | 3329.3 |
| 2022-01-14 00:22:30+00:00 | 0 | 10.75 | 3.5 | 33.09 | 2 | 33.4 | 95 | GU513 | 3329.3 |
| 2022-01-14 00:30:00+00:00 | 0 | 10.75 | 3.5 | 32.29 | 2 | 35.7 | 65 | GU513 | 3329.3 |
| 2022-01-14 00:52:30+00:00 | 0 | 8.9 | 3.58 | 28.19 | 2.03 | 21.8 | 35 | GU513 | 3329.3 |
| 2022-01-14 01:00:00+00:00 | 5.03 | 9.213 | 3.6 | 27.67 | 2.05 | 25 | 85 | GU513 | 3329.3 |
| 2022-01-14 01:07:30+00:00 | 4.95 | 9.013 | 3.55 | 26.76 | 2.1 | 25.1 | 90 | GU513 | 3329.3 |
| 2022-01-14 01:15:00+00:00 | 5.03 | 8.838 | 3.63 | 26.38 | 2.1 | 25.7 | 95 | GU513 | 3329.3 |
| 2022-01-14 01:22:30+00:00 | 5.05 | 8.588 | 3.53 | 26.02 | 2.08 | 24.5 | 90 | GU513 | 3329.3 |
... (164 rows omitted)
day_before_dt.plot('ts', 'TEC')
snap_dt.plot('ts', 'TEC')
The following histograms show first the distribution of TEC values for the day proceeding the eruption, followed second by the same measurement for the day of the eruption. Clearly visible on the day of the eruption is the wide range of values, with highs in the 70 TECUs, compared with 40 and below TECUs the day before.
compare_bins = np.arange(0, 80, 10)
day_before_dt.hist('TEC', bins=compare_bins)
snap_dt.hist('TEC', bins=compare_bins)
plots.clf()
plots.figure(figsize = (20, 10))
plots.plot(
snap_dt.column('ts'),
snap_dt.column('foF2'),
label='foF2',
color='blue'
)
plots.plot(
snap_dt.column('ts'),
snap_dt.column('TEC'),
label='TEC',
color='purple'
)
plots.scatter(x=ERUPTION_START, y=0, color='red', zorder=100)
plots.legend()
plots.show()
<Figure size 432x288 with 0 Axes>
plots.clf()
# We're going to overlay two ionosonde readings,
# so create two subplots:
fig, ax1 = plots.subplots(figsize = (20, 10))
# Instantiate a second axes that shares the same x-axis:
ax2 = ax1.twinx()
ax1.plot(
snap_dt.column('ts'),
snap_dt.column('foF2'),
color='blue'
)
ax2.plot(
snap_dt.column('ts'),
snap_dt.column('TEC'),
color='purple'
)
# Drop a point where the eruption started:
plots.scatter(x=ERUPTION_START, y=0, color="red", zorder=3, label="Eruption start")
ax1.set_title('Digital Ionogram at GU513 Guam')
ax1.set_xlabel('Time (UTC)')
ax1.set_yticks(np.arange(2, 12, step=0.5))
ax1.set_ylabel('Frequency of F2 (MHz)', color='blue')
ax2.set_ylabel('Total Electron Content (10^16 m^-2)', color='purple')
plots.legend()
plots.show()
<Figure size 432x288 with 0 Axes>
# sites = make_array(
# 'AP834', 'E6648', 'F9608', 'C5306', # Hawai'i
# 'AV797', 'AR312', 'AS176', 'C5988', # Californi'a
# 'AP204', 'C6723', 'C7496', 'C8539',
# 'C9585', 'D1970')
# #for site in sites:
# # get_cwop(site)
The block below extracts weather data from the CWOP Data Table within the given timespan and renders it as a line plot.
# event_start = datetime.datetime(2022, 1, 15, tzinfo=datetime.timezone.utc)
# event_end = datetime.datetime(2022, 1, 15, 16, tzinfo=datetime.timezone.utc)
# plots.clf()
# plots.figure(figsize = (20, 10))
# for site in sites:
# cwop_dt = Table().read_table(f"cwop/{site}.csv")
# cwop_dt = Table().with_columns(
# 'ts', cwop_dt.apply(fix_ts, 'Time (UTC)'),
# site, cwop_dt.column('Barometric Pressure (mbar)'),
# )
# cwop_period_dt = cwop_dt.where('ts', are.between(ERUPTION_START, event_end))
# plots.plot(
# cwop_period_dt.column('ts'),
# cwop_period_dt.column(site),
# label=site
# )
# plots.legend()
# plots.show()
time_to_hit_s = 2 * 60 * 60
time_to_hit_s
7200
# 5833872
dist_to_hit_m = 3328.36 * 1000
dist_to_hit_m
3328360.0
dist_to_hit_m / time_to_hit_s
462.27222222222224
# Gets the graphics displayed below, only need to run once:
# !wget https://giro.uml.edu/common/img/giro-map-large.gif
# !wget https://www.swpc.noaa.gov/sites/default/files/styles/medium/public/Ionosphere.gif
# !wget https://www.weather.gov/images/jetstream/atmos/ionlayers.jpg
# !wget https://planetary.s3.amazonaws.com/web/assets/pictures/20170815_ionosonde.png
# !wget https://upload.wikimedia.org/wikipedia/commons/6/69/Ionogramme.png